W.R. Tobin
Professional Website of William R Tobin
Project maintained by wrtobin Hosted on GitHub Pages — Theme by mattgraham
Grad Work
Much of the content below has not been updated since my transition from post-doctoral researcher to research staff at LLNL, but it does still provide an overview of my previous work and research.
Research
My thesis and work was entitled “The Adaptive Multi-scale Simulation Infrastructure (AMSI)”, it was a set of libraries to assist in the development and execution of numerically multi-scale simulations, specifically on HPC machines but targeting in particular the IBM BlueGene/Q architecture. Most performance runs involving AMSI-supported code have been conducted on the AMOS BlueGene/Q machine managed by the CCI.
Additionally I have conducted work to develop numerical HPC simulations and tools including a multi-scale soft biological tissue simulation using the AMSI libraries, the Simmetrix backend for the SCOREC/core tools, and am currently assisting in the development of several fusion codebases:
Libraries and Projects (listed reverse-chronologically)
Where possible the source code repositories are linked, but several projects are only available to the public via request and several projects are not (currently) publicly available due to usage of proprietary APIs from our industry partners. If/when I am able to publicly release API information these repos will be made fully public.
WDMApp, the Whole Device Modeling Application, is a project to couple together different fusion simulation codes to provide a simulation of an whole magnetic confinement-type fusion reactor (such as a Tokamak). The initial implementation couples together the XGC and GENE codebases. Our task is to take the initial implementation which is currently embedded in the GENE codebase and produce a standalone library to support the coupling, as well as the inclusion of additional codes. This project is currently in the planning and design stage, where we are exploring the current implementation and documenting the requirements for our standalone coupling library.
XGCm is a variation on a fusion code developed by the Princeton Particale Physics Laboratory (PPPL). The original XGC code was implemented using structured grids for domain representation, XGCm is being developed by SCOREC to leverage our techniques in unstructured meshing for domain representation. Additionally it is the initial project motivating our work on the PUMIpic library combining unstructured grids and particle-in-cell (PIC) methods and techniques for performing finite element assembly and PIC procedures on GPUs/other accelerators.
m3d-c1 is a fusion code developed by the PPPL. I am currently working on improving the interaction and usage of the code with the meshing and tensor field libraries SCOREC/core and the linear algebraic systems library we’re currently using (PETSc). Primarily I am focused on improving the efficiency of the finite element assembly procedure, w.r.t. both algorithmic and parallel data locality and with restructuring the DOF ordering to give rise to various nonzero patterns in a sparse matrices with a target of improving the iterative linear solver soluation times.
msi is the Mesh Solver Interface. It is intended as a minimal CAPI allowing access to the features in SCOREC/core (PUMI) and their interactions with a linear algebraic system. It is being used to support the implementation of m3d-c1 and XGCm. It is best to think of it as a wrapper layer between various backends, facilitating interactions which require operations from the discretized domain and tensor fields, the linear system, and the numerical kernels supplied by application developers.
LAS is a zero-overhead API for operating on linear algebraic systems operating both in distributed-memory parallel and shared-memory parallel machines. LAS takes advantage of the C++ Curiously Recurring Template Pattern (CRTP) and aggressive inlining to optimize the API function calls away at compile-time. Each linear algebraic system provided to the library at configuration time additionally has a CAPI auto-generated at configuration and compile time which has a single function call overhead to allow using the API from strict C or FORTRAN code since naturally the backend functions cannot be inlined into those languages at compile time.
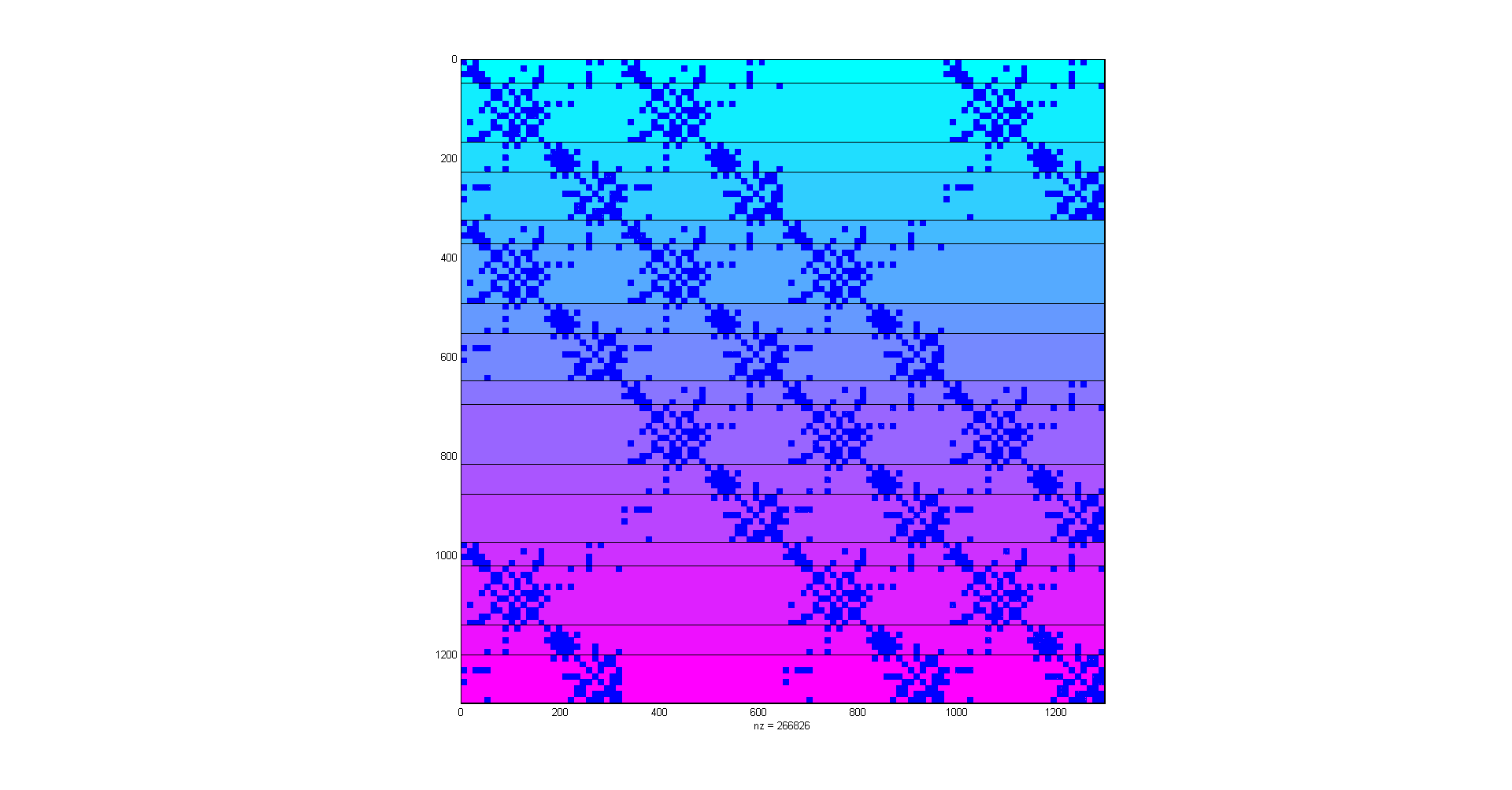
A small PETSc matrix for the M3DC1 simulation using LAS to manage the linear system.
AMSI (contains proprietary API calls from an industry partner so source code is currently private) is a set of libraries used to support the implementation and execution of multi-scale numerical simulations. While many multi-scale systems require adherence to a strict API or set of data structures to which a user must adapt their code, AMSI is intended for use in combining existing well-established single-scale simulation codes. Refactoring and reimplementing a codebase to target a particular multi-scale framework is undesirable, so AMSI requires intervention in exising code only during initialization and at locations where multi-scale values influencing the simulation is required.
Currently AMSI provides the most support for hierarchical multi-scale simulations, where the scales are seperated by many orders of magnitude (typically 6 or more), so inter-domain information required to establish coupling relationships is minimal. Using AMSI in a concurrent multi-scale simulation is certainly possible, but would require the user to implement methods for the interacting scales to establish inter-domain relationships for the scale-coupling. Extending AMSI with generic functionality supporting concurrent multi-scale simulations is a target for future work.
biotissue (also contains API calls from an industry partner) is a multi-scale soft-tissue simulation using AMSI to couple together two scales: an engineering scale (cm and mm scale) and micro-scale (micrometer to nanometer). The engineering scale uses a finite element analysis code based on the Cauchy momentum balance equations for a body in static equilibrium, at every numerical integration point a micro-scale representative volume element (RVE) analysis takes place. Each RVE is a dimensionless bi-unit cube containing a fiber network modeled by truss elements. The macro-scale establishes boundary conditions at the micro-scale RVE corners, which displace the boundary nodes of the fiber network of truss elements. Producing a solution for the micro-scale system gives rise to a set of forces on the boundaries of the RVE which are dimensionalized and sent to the macro-scale for use as stress and stress-derivative terms in the macro-scale governing equations.
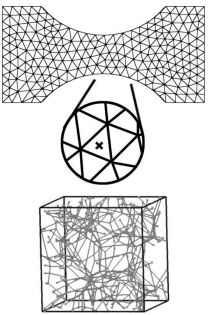
The two biotissue scales: an engineering scale unstructured mesh coupled to dimensionless RVE cube containing a fiber network.
byte_stream is a header-only library using C++11 variadic templates to assist in the serialization and deserialization of data structures. It can handle any plain-old-datatype (POD) trivially, but for datatypes requiring a deep copy it can still provide utility. Mostly it was used to package up small objects to be sent over MPI to avoid defining a custom MPI_Datatype since defining custom MPI_Datatypes is essentially code duplication since you have to define your structure in two places: once in actual code and once to the MPI system.
tensor is a basic implementation of a tensor interface using C++11 variadic templates. It was an attempt at getting a general tensor datatype with low-overhead data access operators to be specialized and used in other libraries needing vectors, matrices, and possibly higher-order tensors, but was discontinued after some initial speculative implementation of a basic dense tensor type (densor).
Scholarly Contributions
Papers / Articles
Chan, Victor W. L. and Tobin, W.R. et al, “Image-Based Multi-scale Mechanical Analysis of Strain Amplification in Neurons Embedded in Collagen Gel”, Computer Methods in Biomechanics and Biomedical Engineering, in submission.
Presentations
*Tobin, W.R. “VS Code in HPC”, IMG Group, March 2023
*Tobin, W.R. “Loop Fusion for Device Packing Kernels”, LLNL ASQ Seminar, March 2021
Tobin, W.R. and Chan, V.W.L. and Shephard, M.S., “Dynamic Load Balancing for Multiscale Simulations”, CSE Seminar Series, May 2015, Rensselear Polytechnic Institute.
Tobin, W.R. and Chan, V.W.L. and Shephard, M.S., “Load Balancing Multiscale Simulations”, SIAM Conference for Computational Science and Engineering 2015, Minisymposium on Partitioning for Multiple Constraints and Objectives.
Tobin, W.R. and Fovargue, D. and Shephard, M.S., “Parallel Infrastructure for Multiscale Simulation”, SIAM Conference for Parallel Processing 2014, Minisymposium on Applications and Impact of FASTMATH II
Awards
Graduate Research Fellowship, Sandia National Labs, 2012-2015
SIAM Travel Award - SIAM Conference for Computational Science and Engineering 2015